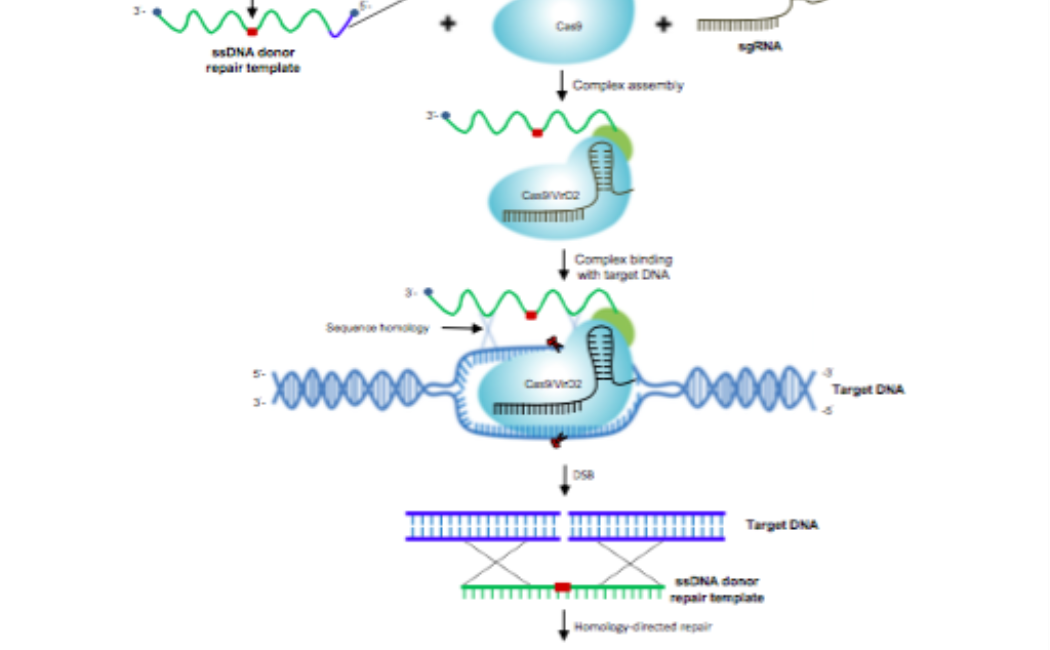
Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice
26 January, 2020
Precise genome editing by systems such as clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) requires high-efficiency homology- directed repair (HDR). Different technologies have been developed to improve HDR but with limited success. Here, we generated a fusion between the Cas9 endonuclease and the Agrobacterium VirD2 relaxase (Cas9-VirD2). This chimeric protein combines the functions of Cas9, which produces targeted and specific DNA double-strand breaks (DSBs), and the VirD2 relaxase, which brings the repair template in close proximity to the DSBs, to facilitate HDR. We successfully employed our Cas9-VirD2 system for precise ACETOLACTATE SYN- THASE (OsALS) allele modification to generate herbicide-resistant rice (Oryza sativa) plants, CAROTENOID CLEAVAGE DIOXYGENASE-7 (OsCCD7) to engineer plant architecture, and generate in-frame fusions with the HA epitope at HISTONE DEACETYLASE (OsHDT) locus. The Cas9-VirD2 system expands our ability to improve agriculturally important traits in crops and opens new possibilities for precision genome engineering across diverse eukaryotic species.